Sample collection
Whole saliva samples were collected using ORAgene®RNA (RE-100, catalog number: RE-100) vial collection kits from DNA Genotek according to the manufacturer’s instructions (DNA Genotek Inc., Kanata, Ontario, Canada). The kit is an all-in-one system for the collection, stabilization and transportation of RNA from saliva. Unstimulated whole saliva was collected from 61 healthy donors (27 females, 34 males, average age 38.5 ± 16.4 years, Fig. 1). The following exclusion criteria were applied: age below 18 years old, donors with history of immunodeficiency, autoimmune disorders, viral hepatitis, HIV infection, current or previous cancer, current oral problems (infection). From all donors, whole saliva was collected from 9 to 11 am and then preserved after having been shaken vigorously in the vial. Additionally, in 10 out of 61 donors, whole saliva samples were collected at 9 am, 3 pm, 9 pm and 9 am again the next day. After completing normal oral hygiene, donors were not allowed to eat or smoke 2 h prior to collection or to drink at least 1 h prior to collection. Samples were stored at room temperature overnight and placed in a freezer (− 20 °C) for storage. All samples were anonymized and obtained with informed consent from the donors. Sampling was carried out in accordance with the institutional guidelines and regulations. At the time of sampling, donors were asked to fill in a questionnaire (supplemental Figure 1) about demographics (e.g. sex, age), social habits (e.g. alcohol, smoking), oral hygiene (e.g. flossing, mouth wash), and previous radiological diagnostic procedures (e.g. number of CT-scans).
RNA extraction
Total RNA comprising a mixture of human and bacterial RNA, was isolated from whole saliva samples following a combination of the ORAgene® RNA purification protocol16 and the mirVana™ kit protocol (Invitrogen™, ThermoFisher Scientific, Carlsbad, CA 92008; USA/Life Technologies, Darmstadt, Germany) as described in detail elsewhere15. In brief, the samples were heated at 50 °C (1 h), three aliquots (of 1000 µl) were generated, incubated at 90 °C (15 min), cooled to room temperature, 40 µl ORAgene® neutralizer solution (1/25 of total volume) was added, incubated on ice, centrifuged at 13,000g (3 min) and the cell-free clear supernatant was collected for further processing. At this step, we switched to the mirVana™ kit protocol17 by adding the Lysis/Binding Solution. With the mirVana™ kit, total RNA, including human and bacterial RNA species, was isolated by combining a Phenol–Chloroform RNA precipitation with further processing using silica membranes. After several washing procedures to purify RNA from other residual debris, DNA residuals were digested on the membrane (RNAse-free DNAse Set, Qiagen, Hilden, Germany). RNA was eluted with 100 µl RNAse free water in a collection tube and the aliquots were pooled for each sample. In order to increase the input RNA amount for downstream gene expression analysis, samples were steamed at 45 °C for 90 min followed by elution with 30 µl of RNase free water before freezing at − 20 °C.
Quality and quantity of isolated total RNA were measured spectrophotometrically using NanoDrop™ One Microvolume UV–Vis spectrophotometer (NanoDrop, PeqLab Biotechnology, Erlangen, Germany). RNA integrity was assessed by the 2100 Agilent Bioanalyzer (Life Science Group, Penzberg, Germany) and DNA contamination was controlled by conventional PCR using actin primers.
Conventional cDNA synthesis—high-capacity cDNA reverse transcription kit
For analyzing gene expression of human rRNA (18S) and pan-bacterial rRNA (16S, see below), total salivary RNA was converted into complementary DNA (cDNA) via reverse transcription using the High-capacity cDNA reverse transcription kit18 (Applied Biosystems™, Life Technologies, Darmstadt, Germany). The amount of total RNA input was always determined to 500 ng measured by NanoDrop™ One. After reverse transcription, cDNA was diluted to a concentration of 0.01 ng/10 µl, which was used for qRT-PCR detection of 16S and 18S rRNA.
Adjustment of human RNA input for cDNA synthesis
A main modification of the previously described workflow15 was the adaption of human RNA input for cDNA synthesis with the SuperScript® III First-Strand Synthesis System (Fig. 2). As NanoDrop™ spectrophotometer only provides total RNA values comprising an unknown mixture of human and bacterial RNA species, we calculated the ratio of the detected raw Ct-values (threshold cycles) of human rRNA (18S) to pan-bacterial rRNA (16S, high-capacity cDNA reverse transcription kit cDNA synthesis followed by 1st qRT-PCR), regarding rRNA as representative for all human and bacterial RNA specimens. The ratio was determined by calculating the relative ratio of 18S rRNA to 16S rRNA for each sample individually as follows:
$$ \textFold\,\textchange = 2^ \wedge (\textCt18\textS\,\textrRNA – \textCt16\textS\,\textrRNA) $$
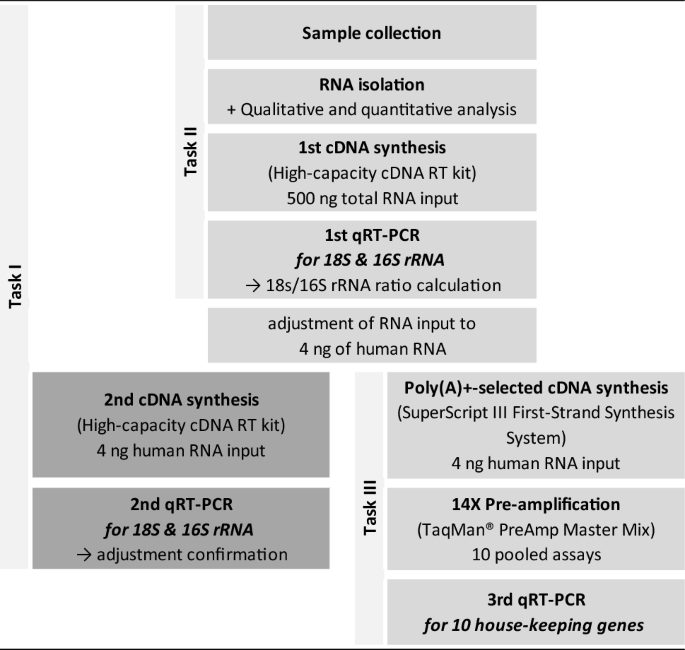
The flow chart displays the different steps (rows) in gene expression analysis including our modified workflow, the tasks, the required kits and the detour for adjustment of human RNA input as well as its confirmation (boxes in darker grey). The boxes in brighter grey depict the advanced methodological workflow for gene expression analysis in whole saliva samples.
Using the generated ratio, a RNA input of 4 ng total human RNA was determined individually for each sample for a second cDNA synthesis followed by 2nd qRT-PCR (confirmation for methodological reasons in this context, Fig. 2 boxes in darker grey).
Modified cDNA synthesis—SuperScript III First-Strand Synthesis System
In order to subsequently perform gene expression analysis of human origin, only eukaryotic messenger RNA (mRNA) was reverse transcribed to cDNA by using the SuperScript® III First-Strand Synthesis System with Oligo (dT)20 primers. As a result, pan-bacterial RNA and other human RNA species were excluded from further processing, minimizing the inhibition effects of those RNA species on the following reactions. According to the kit description, the amount of starting material can vary from 1 pg to 5 μg of RNA and the maximum input volume of RNA is 8 µl19. The amount of human RNA input was set to 4 ng per reaction. Using the concentration from repeated NanoDrop™ measurements and the calculated 18S/16S ratio in each sample, the corresponding amount of measured total RNA input was calculated, conforming to the maximum input volume of 8 µl. The RT was performed according to the manufacturer’s instructions19.
Pre-amplification—TaqMan® PreAmp Master Mix
To detect low abundance mRNA species, pre-amplification was required. We used TaqMan® PreAmp Master Mix (Thermo Fisher Scientific Baltics UAB, Vilnius, Lithuania) to increase the amount of specific cDNA targets, synthesized with the SuperScript® III First-Strand Synthesis System. According to the manufacturer, pre-amplification with this kit is linear when a minimum amount of cDNA molecules is present (minimum of 1–250 ng and Ct-values without pre-amplification should be < 35) and multiplex amplification can be performed by pooling up to 100 TaqMan® Gene Expression Assays. Pre-amplification was performed according to the TaqMan® PreAmp master mix kit protocol20. In the present work, 10 different TaqMan® Gene Expression Assays (ACTB, Hs01060665_g1; B2M, Hs00187842_m1; GUSB, Hs00939627_m1; MT-ATP6, Hs02596862_g1; PGK1, Hs00943178_g1; PP1A, Hs99999904_m1; RPL13A, Hs04194366_g1; RPLP0, Hs02992885_s1; TBP, Hs00427620_m1; YWHAZ, Hs01122445_g1) were utilized and pooled to enable the multiplex amplification of specific cDNA targets. Those are commonly used house-keeping genes already employed in other experimental set ups (www.genomics-online.com).
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Using different sets of primers, two kinds of cDNAs were utilized for qRT-PCR: for human (18S rRNA, Hs99999901_g1) and pan-bacterial (16S rRNA, Ba04230899_s1) primer probe designs, cDNA from High-capacity cDNA reverse transcription kit was used, whereas for the primer probe designs representing the potential house-keeping genes (ACTB, B2M, GUSB, MT-ATP6, PGK1, PP1A, RPL13A, RPLP0, TBP, YWHAZ), SuperScript™ III First-Strand Synthesis SuperMix synthesized, i.e. human cDNA with and without 14× pre-amplification was used for detection of each of these genes in each sample. The qRT-PCR reaction contained TaqMan® Universal PCR Master Mix and one of the inventoried TaqMan® Gene Expression Assays for separate detection of transcripts. All measurements were run in duplicate, using a 96-well-format TaqMan® qRT-PCR platform and the QuantStudio™ 12 K Flex Real-Time PCR System.
Statistical analysis
Descriptive statistics (n, mean, standard deviation, minimum [min], maximum [max]) were calculated for continuous variables such as gene expression data and age. Frequency tables of categorical data were examined for statistical differences using the chi-square test for equal proportion. Comparisons of categorical variables with gene expression values were performed using the non-parametric Kruskal–Wallis (KW) test. We assessed the assumptions of normality (Kolmogorov–Smirnov) and boxcox transformed the continuous variable “age”. All calculations were performed using SAS (release 9.4, Cary NC, USA). Graphical presentations were performed using Sigma Plot 14 (Jandel Scientific, Erkrath, Germany).
Data availability and approval statement
The merged set of raw data is provided within supplemental Table 2. The qRT-PCR related measurements (e.g. RNA quantity/quality and TaqMan® qRT-PCR) were performed according to the standard operating procedures implemented in our laboratory in 2008 when the Bundeswehr Institute of Radiobiology became DIN-certified by TÜV Süd München, Germany (DIN EN ISO 9001/2008). All samples and data was processed anonymously without exception and only for this specific purpose. All data is handled according the European General Data Protection Regulation. Data will be deleted after 10 years. Due to the minimal-invasive collection and the fully anonymized processing of the samples, the local ethical commission (Ethics committee, Bayerische Landesärztekammer, Munich, Germany) decided that experiments can be performed in agreement with ethical standards and do not require an additional approval.





More Stories
Must-Have Housekeeping Supplies for Every Household
Top Natural Cleaning Products for a Healthier Home
The Ultimate Housekeeping Checklist for Every Home